Workshop on Reproducible Research
A hands-on introduction to quarto with a Computo submission
Planning
- 9h-10h: Introduction to Computo and Quarto
- 10h-10h30: Coffee break/Discussion
- 10h30-11h30: Hands-on with a toy example
- 11h30-12h30: Follow-up and optional personal article submission
Learning objectives
- Understand the benefits of reproducible research
- Learn how to create a quarto document
- Learn how to include code, data, and narrative text in a quarto document
- Learn how to submit a quarto document to Computo
- How to navigate the Computo submission process (optional)
Short introduction to Computo and quarto
Team
Editorial board
IT support

Julien Chiquet (chief editor)
Stat. learning, DR INRAE
Paris-Saclay University

Pierre Neuvial
Statistique, DR CNRS
IMT Toulouse

Mathurin Massias
Optim./Machine-Learning
CR INRIA Lyon

Fra.-Dav. Collin
CS/Stats/ML, IR CNRS
IMAG, Montpellier University

Nelle Varoquaux
Machine learning, CR CNRS
Grenoble Alpes University

Marie-Pierre Étienne
Statistics, MCF
Institut Agro Rennes-Angers

Chloé Azencott
Machine Learning
CR MinesParisTech

Ghislain Durif
Stats/ML/dev, IR CNRS
LBMC, ENS LYON
What is reproducible research?
Fundamentally, it provides three things:
Tools to reproduce the results (that’s like cooking)
A “recipe” to reproduce the results (still like cooking)
A path to understand the results and the process that led to them (unlike cooking…1)
Pre-Computo era
The pdf era and paper submission.
The reproducibility was not a priority:
- Tools has to be bought, installed, and maintained
- Data and code were not shared (social engineering
- Even methodology details are often missing

Social engineering required to get reproducible results: at best you just have to ask the authors, at worst you have to reverse-engineer everything… and have no guarantee whatsoever to get to the same results, when authors are in a “Après moi, le déluge” mood and ditch, forget or let it crumble once the paper is published.

And then in the Machine Learning domain, there was distill.pub [1]
- state-of-the art visualizations
- paradigm shift in the scientific publication: “distillation” of complex ideas
- 100% reproducible (just a git clone and a few standard commands)
but…

… engineering was too complex for the average scientist (a lot of javascript, etc.)
In fact, the distill.pub project was discontinued in 2021 [2]
Due to a serie of burnouts from the staff
The Rise of the Pragmatic
distill.pub’s goals were right, but they outpaced themselves in terms of development complexity.
- Computo is a fresh start with a pragmatic approach
- leverage what the scientific community is already using (Rmarkdown, Jupyter notebooks, etc.)
Origin of Computo (~ 2020s)
French statistical society appoints a “publication” committee (lead by Julien then Pierre) to develop a new journal
- 😔 Multiplication of “traditional” journals…
- 😔 No valorization of “negative” results
- 😥 No or not enough valorization of source codes and case studies
- 😱 ↘ of publication quality and time dedicated to each article (on author or reviewer sides) [3]
- 😱 Issue with scientific reproducibility (analyses, experiments) [4–9]
- Need for renewal regarding scientific research implementation
- Need for higher standard regarding result publications
⇝ Emergence of “Computo” idea
Philosophy
Promote contribution in statistics and machine learning that provide insight into which models or methods are more appropriate to address a specific scientific question
- “Diamond” open access (free to publish and free to read, possible to reuse)
- 🅭 🅯 Content published under CC-BY license (attribution, share, adapt)
- Reviews and discussions available after acceptance for publication (anonymous reviews)
⇝ In accordance with Budapest Open Access Initiative (BOAI) and Plan S

- Numerical (statistical) reproducibility is a necessary condition
- Source code and data should be available, at least partly executed and fully executable
Note on reproducible research [10–12]
Why reproducing scientific results?
- To strengthen their credibility
- To check for errors (everyone makes error at some point!!!)
- To build new research upon them (science is incremental)
Issues?
- Reproduce numerical scientific results is often difficult (technology/environment evolution, source code/environment configuration/software partially available or not available)
- Waste of time and resources to reproduce existing non-reproducible results
Reproducible research?
- For others but also for your future self
- Improve result credibility
- Facilitate future research works
Setup
Official launch at the end of 2021
“Economical” model
- A few tenacious people…
- Free/Open-source community tools (Pandoc, Quarto, Git forge)
- Institutional support (INRAE, INRIA, CNRS, SFdS)
Functioning
Writing system
Notebook and literate programming
text (markdown) + math (\(\LaTeX\)) + code (Python/R/Julia), references (bib\(\TeX\))
Publication system
Environment management, Compilation, Multi-format publication (pdf, html)
Continuous integration/Continuous deployment (CI/CD)
Reviewing system
- Anonymous exchange published after acceptance
- Reviewer pool (you can join)
- [Ongoing switch from Scholastica to Open review]
Note on literate programming
- Literate programming [13]: notebook including text and code
- Markup formatting language: e.g.
markdown - Separate content from rendering (≠ “what you see is what you get” editors)
- Rendering includes text, code and results (from code computations)
---
title: "My article"
---
We compute 1+1:
\`\`\`{r}
1+1
\`\`\`Note on quarto
- Generalization of
Rmarkdown - Relying on top community tools like universal document converter
Pandoc - Developed and supported by RStudio/Posit
- Native support of complex documents (website, articles, books) and multiple languages for computations (R, Python, Julia)
- Management of references, citations, figures, tables, metadata, etc.
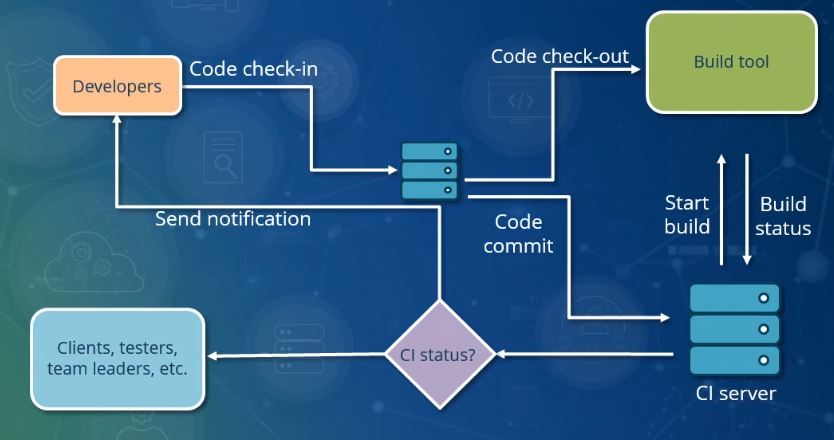
Note on continuous integration
- Implementation in git forges (e.g. github actions or gitlab CI/CD)
- Triggered by commits
- Automatic tests
- Automatic deployment: package/software publication, website
Credit: Pratik89Roy CC-BY-SA-4.0 from Wikimedia
Editor point of view
After a “traditionnal” review process, a 3 step procedure:
- Acceptance
- Pre-production
- Publication in Computo (with a DOI)
including
- Copy of the author git repository to https://github.com/computorg/
- Final version formatting
- Review report publication
- Registration in the journal bibliographic data base
- Copy of the repository to Software Heritage for archiving
- Publication of the article on the journal website
2year and a half report
🥲 Fully operational + doi, ISSN
🙂 7 published articles articles, 3 in preproduction, 6 under review (more details here)
🙂 x presentations (Montpellier, Toronto, Humastica, Grenoble, RR2023, etc.)
🙂 French reproducible research network
🤯 Difficult to find reviewers
🤔 Institutional support?
🤔 Changing of practices in the scientific community?
Discussion
About several choices
Comparison/inspiration
- Peer Community-In (PCI)6, EpiSciences: different philosophy and/or functioning
- https://rescience.github.io/: “remake” published articles
- https://distill.pub (discontinued): technically more complicated and only ML/AI oriented
Perspectives
- Provision of computing resources (to be able to run all computations)
- Full gitlab support (CI/CD, docker, registry, etc.)
- Switch to a french institutional gitlab forge?
- Improve long-term reproducibility stack (docker container, GUIX fully reproducible environment, only at the end of the publication process, )
How to help?
Regarding the logo
References
Reproducibility considerations
Two-fold reproducibility
The global scientific workflow of a reproducible process for a Computo may be split in two types of steps:
External and Editorial
External
- External
- Process to obtain (intermediate) results utside of the notebook environment, for a list of reasons (non-exclusive to each other):
- the process is too long to be conducted in a notebook
- the data to be processed is too big to be handled directly in the notebook
- it needs a specific environment (e.g. a cluster, with gpus, etc.)
- it needs to involve specific languages (e.g. C, C++, Fortran, etc.) or build tools (e.g. make, cmake, etc.)
Editorial
- Editorial
- notebook rendering with the results of the external process
If the notebook contains everything to produce the final document
\(\Rightarrow\) “Direct reproducibility” in the sense that the notebook is the only thing needed to reproduce the results.
Ultimately, the workflow must end with a direct reproducibility step which concludes the whole process.
- Data transfer
- When applicable, the switch from external to editorial reproducibility is done with a “data transfer” step,
data produced by the external process \(\Rightarrow\) transferred to the notebook environment.
Not only the intermediate results are provided, but also the code to transfer it in the notebook environment.
They are a variety of software solutions to do so.
Examples of data transfer solutions
Intermediate results storage
- Python:
joblib.Memory, caching mechanism for python functions, save the results of a function call to disk, and load it back later. - R :
.RDatafile format, can be loaded back in R with theload()function. - If results are small enough, storing in a text file (e.g.
.csv,.tsv,.json, etc.) is also a solution.
Transfer of the results to the notebook environment
- (
.joblibdirectory or.Rdatafile) could be committed to the git repository, and directly loaded in the notebook environment. - Alternative, centralize input data (when large enough) and intermediate results on a shared scientific provider (we recommend Zenodo for this purpose), and download them in the notebook environment.
Workshop
Quarto
In this workshop, we will learn how to use quarto to create a document that includes code, data, and narrative text. We will also learn how to make the CI (continuous integration) working.
The main pipeline, step by step
- Template installation
- computing environment : renv, conda, etc.
- Authoring in the qmd
- rendering locally
- pushing to github
Getting started
To get started you will need to clone the mock template for this workshop. The template is available at
https://github.com/computorg/template-jds2024
Mock repository
https://github.com/computorg/template-jds2024
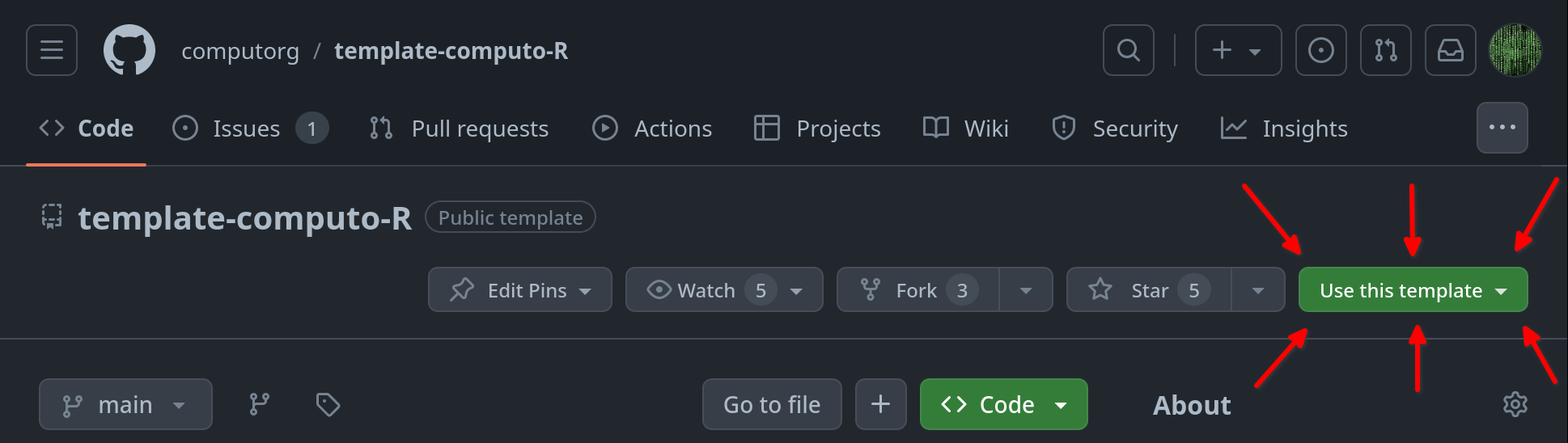
Creating a repo from a template
- On GitHub.com, navigate to the main page of the repository.
- Above the file list, click Use this template.
- Select Create a new repository.
- Select
Include all branches.
Language version
Make a git clone of the repository you just templated and open it in your favorite IDE.
Python version
Rename the published-paper-tsne-python.qmd to published-paper-tsne.qmd
R version
Rename the published-paper-tsne-R.qmd to published-paper-tsne.qmd
Conclusion
Footnotes
Even so, we may discuss the fact that blindly following recipes will not make you a good cook.↩︎
with CI/CD support↩︎
or as a gitlab page when gitlab will be supported (soon)↩︎
and soon the
.gitlab-ci.ymlfile for the gitlab CI/CD configuration↩︎“free and open-source”↩︎
Computo is a PCI-friendly journal↩︎
contact us at computo@sfds.asso.fr↩︎